
Lorincz Lab
Our lab studies the role of histone modifications, DNA methylation, and chromatin remodelling factors in transcriptional regulation of genes and retroelements during mouse development using next-generation sequencing technologies.
We are located at the Life Science Institute, University of British Columbia, Vancouver, Canada.
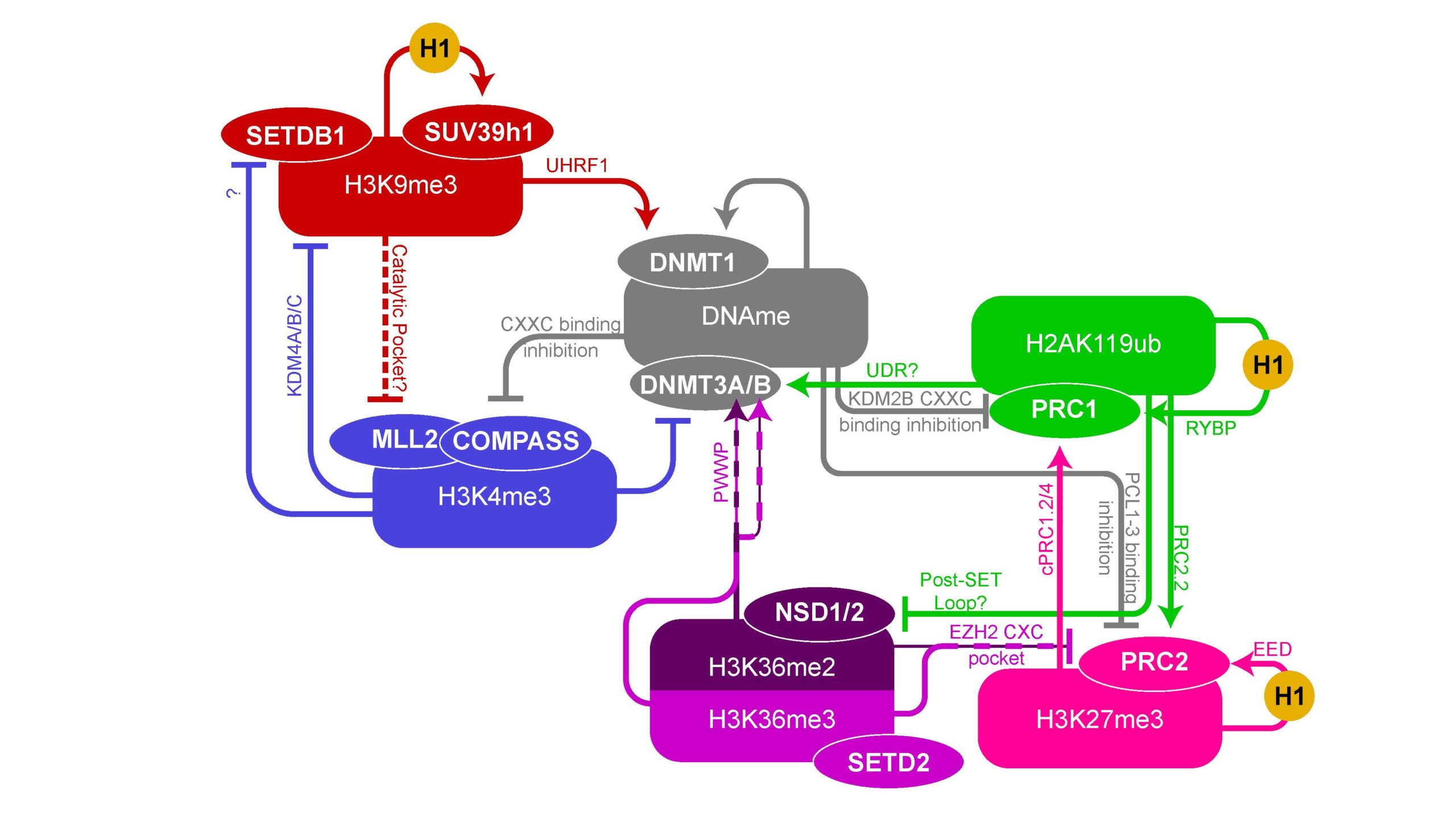
Chromatin marks feature diverse interactions, sometimes promoting the acquisition of new marks and other times precluding them. DNA methylation is a central hub and theme of the research of our lab, with many interactions through various histone marks.
see Publications or Scopus for complete list of publications from the Lorincz Lab

